3. Sampling the Imaginary
Easy.
The code used to answer the questions below relies on these Python libraries:
import numpy as np
import scipy.stats as stats
import matplotlib.pyplot as plt
import arviz as az
# I'm using this code to make plotting easy in a Jupyter notebook
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
plt.style.use(['seaborn-colorblind', 'seaborn-darkgrid'])As in Chapter 2, we’ll compute the grid approximation of the posterior distribution using this helper function written by the wonderful people at PyMC3:
def compute_grid_approximation(prior, success=6, tosses=9):
"""
Parameters:
prior: np.array
A distribution representing our state of knowledge
before seeing the data. The number of items
should be the same as the number of grid points.
success: integer
Number of successes.
The default is 6.
tosses: integer
Number of tosses (i.e. successes + failures).
The default is 9.
Returns:
p_grid: np.array
An evenly-spaced parameter grid between 0 and 1.
posterior: np.array
The posterior distribution.
"""
# First, define the parameter grid
p_grid = np.linspace(0, 1, prior.shape[0])
# Then compute the likelihood at each point in the grid
likelihood = stats.binom.pmf(success, tosses, p_grid)
# Compute the product of the likelihood and the prior
unstd_posterior = likelihood * prior
# Finally, standardize the posterior so that it sums to 1
posterior = unstd_posterior / unstd_posterior.sum()
return p_grid, posterior, success, tossesWe’ll also use this helper function to visualize the results:
def plot_interval(samples, left=0, right=1, xvar=None):
fig, ax = plt.subplots()
ax.axvspan(left, right, facecolor='grey', alpha=0.35)
ax.hist(samples, bins=100)
ax.set_xlim(0, 1)
yt = list((str(int(i)) for i in ax.get_yticks()))
yt[0] = None
ax.set_yticklabels(yt)
plt.xlabel(f'Probability of {xvar}')
plt.ylabel('Frequency')The questions below rely on a set of samples specified in the R code 3.27 box found on p. 69 of Statistical Rethinking. We’ll generate the samples using the following code:
var = 'Water'
size = 1000
# Create a prior distribution of 1000 1's
# This will also create a posterior distribution with 1000 points
prior = np.ones(size)
pg, po, s, t = compute_grid_approximation(prior, success=6, tosses=9)
# Take 1000 samples (with replacement) from the posterior
# The posterior provides the probability for each value in the parameter grid
# Setting a random seed allows the results to be reproducible
np.random.seed(3)
samples = np.random.choice(pg, p=po, size=size, replace=True)
plot_interval(samples, right=0, xvar=var)Documentation for np.random.seed
Documentation for np.random.choice
3E1. How much posterior probability lies below p = 0.2?
np.mean(samples < 0.2)
plot_interval(samples, right=0.2, xvar=var)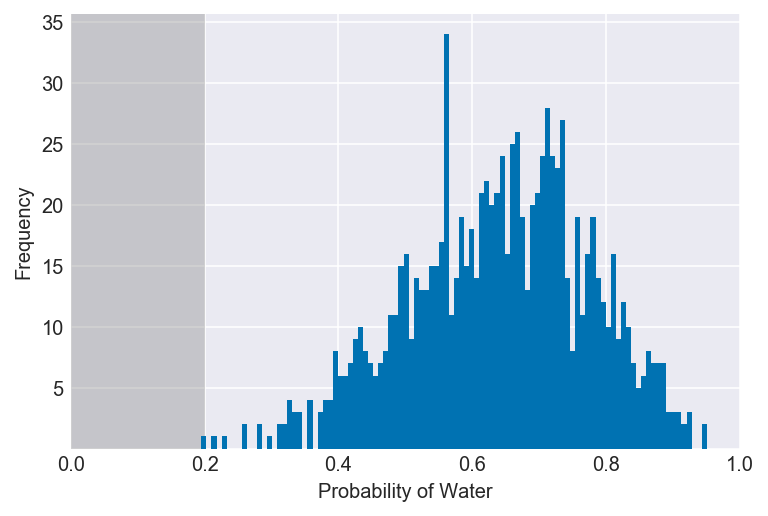
3E2. How much posterior probability lies above p = 0.8?
np.mean(samples > 0.8)
plot_interval(samples, left=0.8, xvar=var)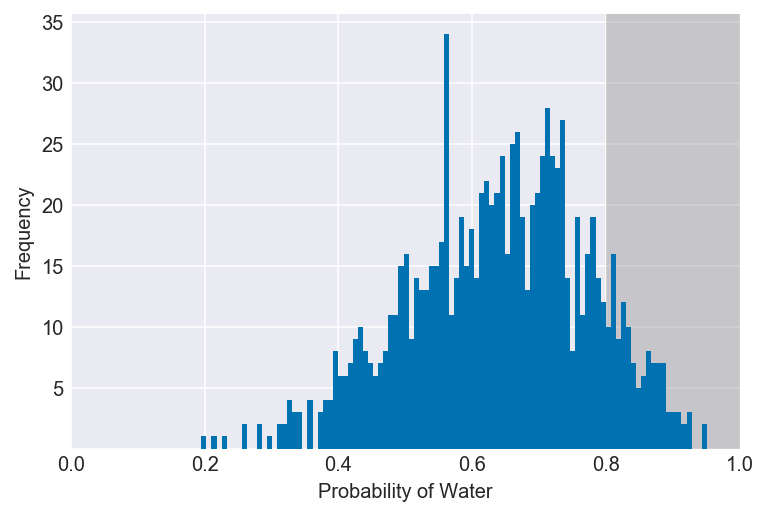
3E3. How much posterior probability lies between p = 0.2 and p = 0.8?
np.mean((samples > 0.2) & (samples < 0.8))
plot_interval(samples, left=0.2, right=0.8, xvar=var)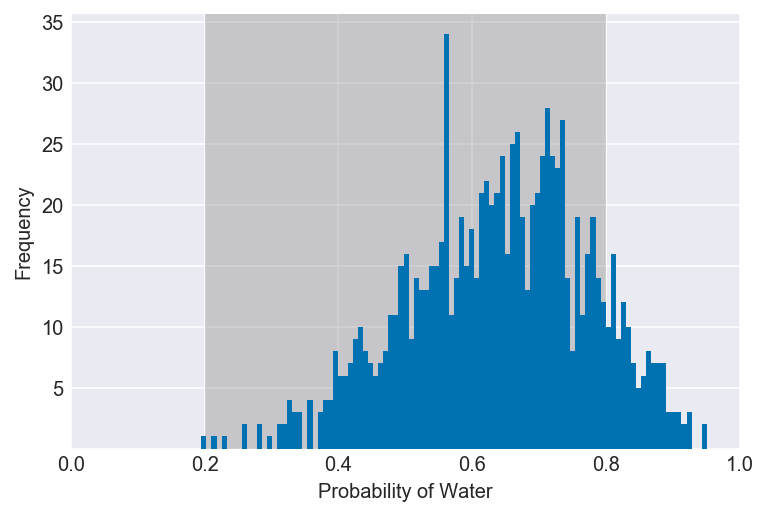
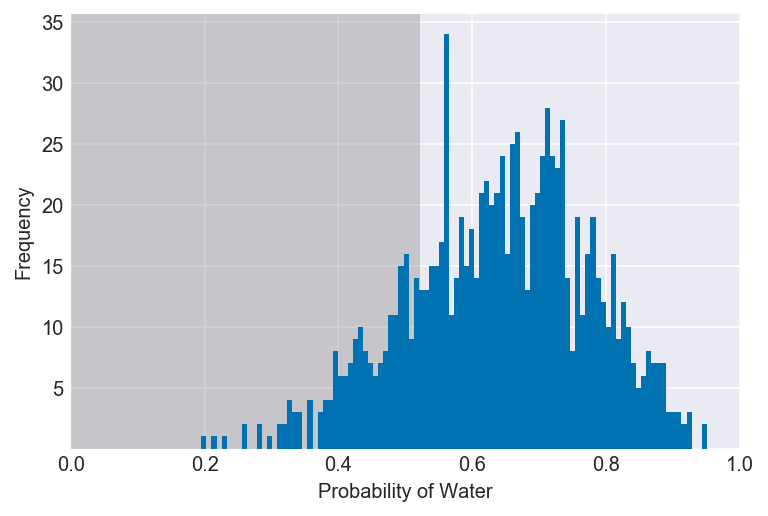
3E4. 20% of the posterior probability lies below which value of p?
value = np.percentile(samples, 20)
plot_interval(samples, right=value, xvar=var)Documentation for np.percentile
3E5. 20% of the posterior probability lies above which value of p?
This is equivalent to asking the value of p that lies below 80%.
value = np.percentile(samples, 100-20)
plot_interval(samples, left=value, xvar=var)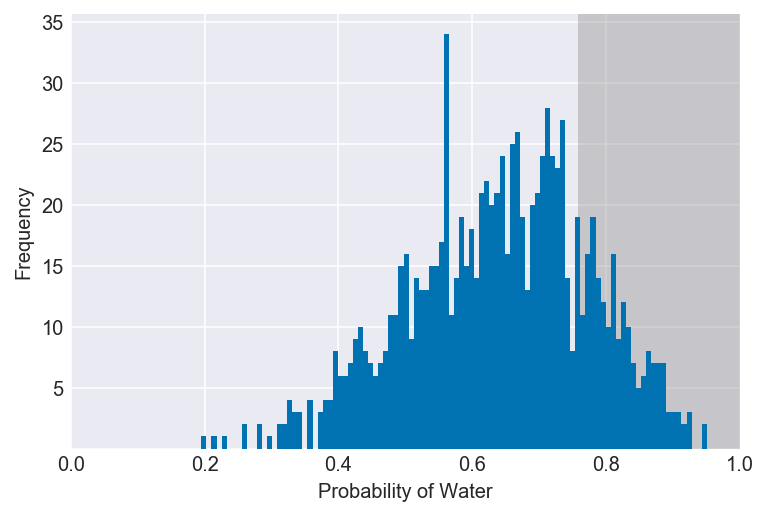
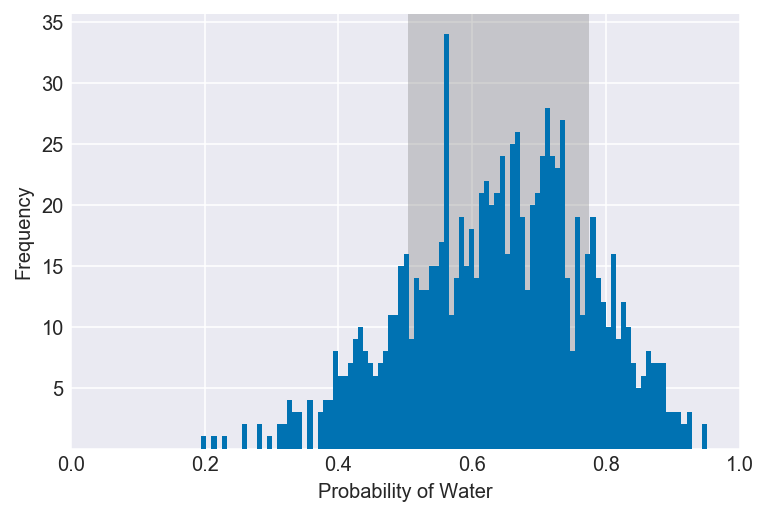
3E6. Which values of p contain the narrowest interval equal to 66% of the posterior probability?
values = az.hdi(samples, hdi_prob=0.66)
plot_interval(samples, left=values[0], right=values[1], xvar=var)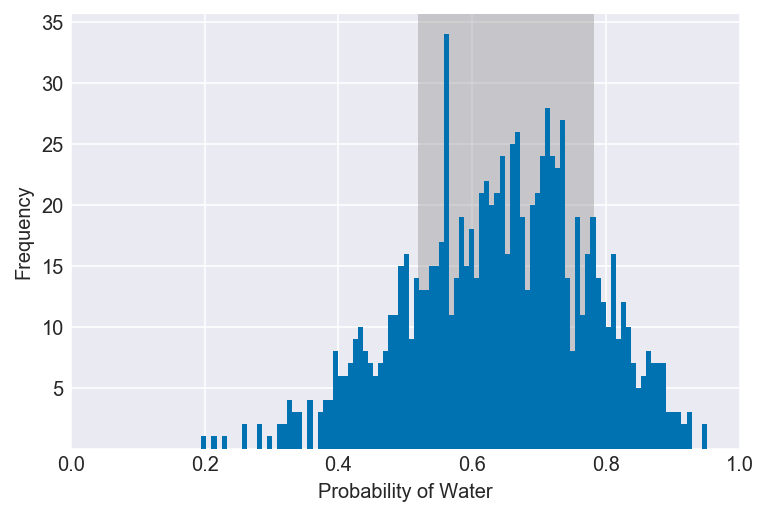
3E7. Which values of p contain 66% of the posterior probability, assuming equal posterior probability both below and above the interval?
If there is an equal posterior probability both below and above the interval, then the remaining intervals comprise the lowest and highest 17% of the posterior (17 + 66 + 17 = 100). We need to compute the interval between those two percentiles.
values = np.percentile(samples, [17, 100-17])
plot_interval(samples, left=values[0], right=values[1], xvar=var)